
Life Science

INDUSTRY
Life Science
Ključna riječ
Uvođenje usluga i proizvoda
CELL PICKER Automated Pick and Collection Tool of Cell Colonies
Associate Professor Iimori is pursuing research based on “Ikuyaku” (drug fostering and evolution).” The intent is to understand antitumor effects in order to further enhance them by conducting a detailed analysis of antitumor drugs with mechanisms of action that are not fully understood but that are already in clinical use. We asked him how he learned about CELL PICKER in the course of pursuing cooperative research with pharmaceutical companies and surgical classrooms in drug fostering and evolution. We also asked him how he uses the instrument.

Associate Professor, Makoto Iimori
Department of Molecular Cancer Biology, Graduate School of Pharmaceutical Sciences, Kyushu University
Note: The details posted here (organizational affiliations and job titles) were accurate at the time of the article. Kyushu University Website
*Podružnice i naslovi ispitanika su aktualni od vremena izvještavanja.
Kyushu University
Associate Professor Iimori and CELL PICKER
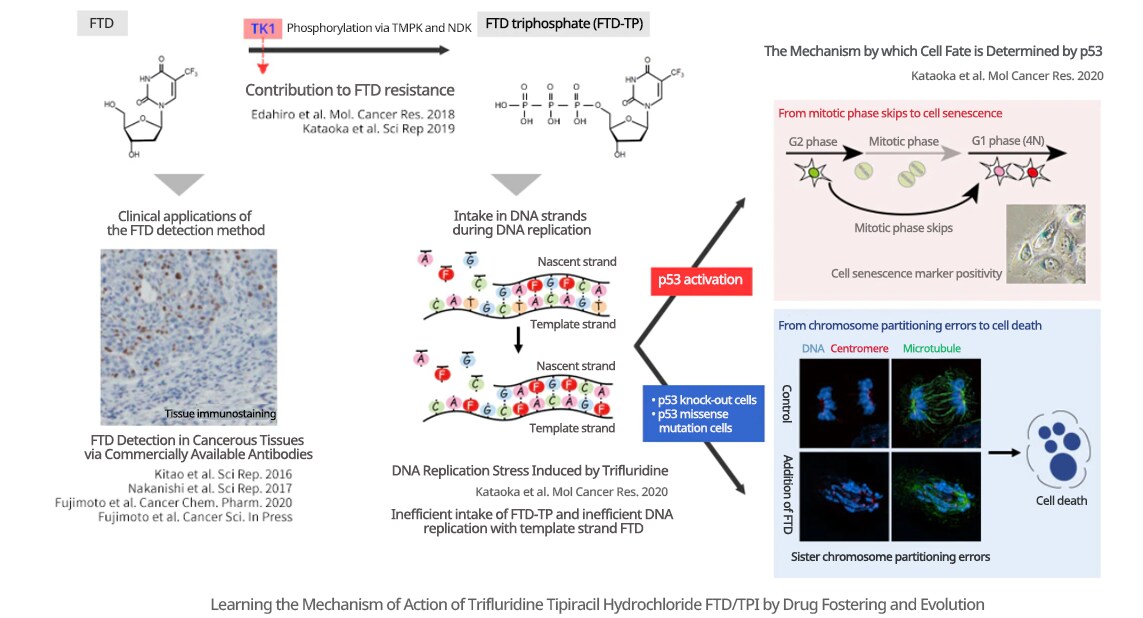
I am pursuing research regarding why antitumor drugs are effective, and by what mechanism they are related to resistance. My basic research aims to further improve the therapeutic performance of antitumor drugs with adjuvant chemotherapy. As an example, I am observing and reviewing the effects of Lonsurf (FTD/TPI), an antitumor drug already in use for colorectal cancer and stomach cancer (see the figure below). When it was first released, it was known that the intake of FTD into chromosomal DNA during the DNA replication cell cycle was necessary for its antitumor effects. However, this mechanism of action was insufficiently understood. Accordingly, by observing the cellular response induced by FTD, we were able to clarify that when FTD is taken into DNA, it decreases the efficiency of DNA replication, thereby causing DNA replication stress. Further, we discovered that DNA replication stress-activated p53-p21, inducing mitotic phase skips, and subsequent cellular senescence, and other cell arrests. In addition, with p53 knock-out cells and missense mutation knock-in cells created using genome editing, a transition to the mitotic phase was observed after DNA replication stress, as was the induction of large-scale chromosome partitioning errors and subsequent cell death. We are also taking the approach of looking at clinical applications, clarifying the contribution of the expression level of thymidine kinase 1 (TK1) to FTD resistance, and developing methods of visualizing FTD in cancer tissues.

Research Laboratory of Professor Hiroyuki Kitao
Genome editing techniques have become commonplace since the release of the CRISPR/Cas9 system. In our laboratory, we were increasingly presented with opportunities to cultivate knock-out or knock-in cell strains. Further, as far as novices are concerned, there were limits in terms of time and technique on the rate of isolating the colonies required when creating cell strains. In addition, even experienced analysts found the process of isolating colonies a time-consuming burden.
With this in mind, I learned of CELL PICKER at a Shimadzu booth that I visited at an academic exhibition. Further, I was given an opportunity to examine the actual instrument, and it roused my interest.
To that point, when creating cell strains with stable expression of genes of interest, as well as gene knock-in and knock-out cell strains, I had isolated colonies using the cloning ring method and the limiting dilution method. A degree of technical expertise is required to use the cloning ring method. In addition, it is necessary to pay extremely close attention to the colony formation density in order to deploy a ring so as to enclose the colony to isolate. The colony formation density depends on the cell strain type and the efficiency of plasmid introduction to the cells. Accordingly, cells are seeded following gene transfer under multiple dilution ratio conditions, which requires a number of dishes. In contrast, with CELL PICKER, since the colony is isolated directly, I presumed that there was basically no need to pay attention to the colony density. The limiting dilution method requires even more time and effort, and I felt that there were limits to how quickly the cell isolation process could proceed.

Process of Isolating Cell Colonies
Firstly, it was clear that the instrument was compact, and that it could be operated easily within a clean bench. In addition, the pickup motion was smooth, and there was no sense of time pressure. Further, an image of the cells in the dish is visible on a tablet while proceeding, so there is no need for analysts to peer into a microscope. This meant that everything could be done with the clean bench aperture minimized, which I felt was advantageous from the perspective of working under sterile conditions.
What led me to purchase the instrument was the fact that all of the laboratory personnel who operated the instrument during the demonstration were able to isolate cells without special training, regardless of individual differences in their skill at isolating cell colonies (everyone from people who routinely isolate cells using the cloning ring process to those who had never seen the cloning process itself). This means that not only experienced analysts but also students affiliated with the laboratory could immediately perform this operation. Because it takes one to two weeks to form a colony with cell cloning, quick, convenient, and reliable colony isolation techniques are also important to avoid wasting research time and money.
We routinely use it to establish knock-out or knock-in cell strains using the CRISPR/Cas9 genome editing system technology. Before that time, our experience was with gene knock-out and point mutation knock-in, as well as knock-ins of GFP and other fluorescent proteins and tag sequences. Recently, time-specific protein degradation via auxin-inducible degron, a method developed by Professor Masato Kanemaki’s group from the National Institute for Genetics, has become indispensable in our laboratory. In such cases, knock-in is performed in order to add mAID tags to genes of interest using the CRISPR/Cas9 system. In addition, even when strains with stable expression of a gene of interest were obtained, to that point, the genomes were randomly integrated. Recently, however, using the CRISPR/Cas9 system, we cut the safe harbor locus AAVS1, and integrate the gene expressing plasmid there. After obtaining cells with the intended genotype via the above-mentioned process, we remove cassettes responsible for resistance to the pharmaceutical agent that are fitted into the loxP sequence by Cre Recombinase, and by further integrating the necessary reporter genes, establish the cell strain required for the intended experiment. Cell colony isolation is necessary at each step, so CELL PICKER is always required while creating the cells. (See the figure to the right.)
In terms of technical tips, the colonies that have been picked up are placed temporarily in a cell dissociation solution. The solution used in this case is not a trypsin solution, but rather a TrypLE reagent (from Thermo). With TrypLE reagent, favorable cell adhesion has been confirmed even after cells being picked up from 48 colonies are immersed one after the other in the cell dissociation solution.

Creating Knock-in Cells by Adding a Tag (such as GFP or FLAG) to the End of a Gene of Interest
Professor Kitao (third from left), Associate Professor Iimori (second from left), and the Other Members of the Laboratory
At present, tip attachment and positioning of the 96 well dish when seeding are performed manually. However, I have heard that an automated instrument is currently in development, and I am very much looking forward to that. In addition, with regards to pick up, it is evident that the manner of pickup differs with the type of cell. For example, with some cells, the colony is partially destroyed by pickup, while for other cells, the adhesion between cells when the colony is formed is strong, so the colony as whole peels away as a single sheet. Increasing the variation in pickup movements would likely further increase the efficiency of pickup with respect to these differences.
CELL PICKER, which is described here, has not been certified or validated as a medical device under the Japanese Pharmaceutical and Medical Device Act. It cannot be used for diagnostic treatment or such processes.